Introduction
The oral cavity is a unique ecosystem in which teeth, gingival groove, tongue, cheeks, soft and hard palate, and tonsils coexist to create a special environment. It is covered with mucosa, which consists of keratinized or nonkeratinized squamous stratified epithelium, depending on the localization and function. The oral cavity, being an extension of the surrounding external environment, is colonized by billions of bacteria, viruses, and fungi that form the microbiome of the oral cavity. This commensal microbiota coexists in symbiosis with its host, but when an imbalance occurs, the disease develops. This imbalance is called dysbiosis. Dental caries and periodontal diseases, the main dental conditions, are caused by dysbiosis. In addition to the commensal flora in the oral cavity, there also occurs a transient flora, e.g. the intestinal flora, which resides in the oral cavity for a short period of time due to difficulty in acquiring nutrients [1, 2].
The microbiota of the oral cavity is also a risk factor for many systemic diseases such as tumours [3], diabetes [4], cardiovascular diseases [5], sepsis [6], preterm births, and low birth weight of infants [7]. More than 700 species of bacteria were identified with culture-independent approaches, and more than 250 were isolated, cultured, and named [8]. Undoubtedly, new species are still being identified [9, 10].
Salivary microbial
The greatest number and diversity of microbiota are observed in dental plaque, then in saliva, and on the oral mucosa. The dominant fraction of microbiota in the saliva is like that found in the throat and tonsils. In addition, saliva also contains bacteria from supra and subgingival niches. The main types of bacteria noticed in saliva include Actinobacteria, Bacteroidetes, Firmicutes, Fusobacteria, and Proteobacteria [11]. It has been proven that the salivary microbial configuration is more dependent on the influence of the environment than on genetic conditions [12].
Dental plaque
Dental plaque is a type of biofilm located on the surface of the teeth. There are 2 plaque classifications: supragingival plaque (placed above the gingival margin) and subgingival plaque (below the gingival margin). The composition of the subgingival plaque depends on the balance between health and disease and is associated with periodontal destruction. Microbiological research of this niche focuses mainly on changes in the microbiota that occur during the development of periodontal diseases. In gingivitis, which is an inflammatory process confined to the gum and most often associated with the accumulation of plaque, the dominant microorganisms are: Streptococcus, Actinomycetes, Capnocytophaga, Campylobacter, Eikenella, Fusobacterium, and Prevotella. On the other hand, in the case of periodontitis, which is a chronic inflammatory process involving not only the destruction of the gingiva, but also tooth-supporting tissues (periodontal membrane and alveolar bone), the subgingival microbiota in the course of periodontitis is very complex. Over 400 phenotypes have been isolated from periodontal pockets [13].
Oral mucosa
Compared with the other parts of the oral cavity, microorganisms existing on the mucosa are few. The dorsum of the tongue is the most colonized, by bacteria associated with halitosis. Among the bacteria on its surface in healthy populations, we can identify Streptococcus salivarius, Rothia mucilaginosa, and unidentified, cultivable species of Eubacterium strain FTB41 [13].
Diabetes mellitus
Various research studies on the composition of the subgingival bacterial microbiota in diabetics compared to healthy individuals have been performed. However, no clear influence of diabetes on bacterial flora has been demonstrated.
Certain species of bacteria are more common in diabetic patients. However, it is difficult to confirm whether their occurrence is directly related to a change in the subgingival environment or indirectly due to a change in the host’s immune response. Diabetic individuals are more prone to chronic periodontitis as a result of hyperglycaemia that alters the subgingival environment such that pathogenic bacteria become dominant. Alternatively, diabetes can alter the host response to plaque, causing more tissue destruction. Considering the elevated glucose level in the subgingival environment and the altered or inhibited host immune response, real differences can be found in the subgingival microbiome in diabetic patients in comparison to healthy individuals [14].
Aim of the study
The aim of the study was to analyse and compare the composition of bacterial oral microbiota in children with type 1 diabetes and healthy children.
Material and methods
Characteristics of the research groups
The study group consisted of 50 randomly selected children aged 10–18 years, who had had type 1 diabetes for at least 5 years. The subjects were patients of the regional diabetic clinic at the Upper John Paul II Silesian Children’s Health Centre University Hospital in Katowice, Poland.
Patients from the study group were divided into 2 groups (25 people each) according to their level of diabetes control. The allocation of patients to the groups was based on the qualification criterion, which was the level of glycated haemoglobin (HbA1c). According to the recommendations of the American Diabetes Association (ADA) and the International Society for Paediatric and Adolescent Diabetes (ISPAD 2014), the threshold value of HbA1c was set at 7.5% [15]. Patients with good glycaemic control were assigned to the Well Controlled group (WC, HbA1c ≤ 7.5%). The mean age of the subjects in the WC group was 14.40 ±2.0 years. The group consisted of 10 girls and 15 boys. Twenty-three patients in this group were using a personal insulin pumps (continuous subcutaneous insulin infusion-CSII) and 2 patients were using insulin pens (multiple daily insulin injection-MDII). The average level of glycated haemoglobin was 6.70 ±0.40%. The Poorly Controlled (PC, HbA1c > 7.5%) group consisted of patients with poor glycaemic control. The mean age of the subjects in the PC group was 14.92 ±1.87 years. The group consisted of 14 girls and 11 boys. Eighteen patients in this group were using personal insulin pumps (CSII) and 7 were using insulin pens (MDII). The mean glycated haemoglobin level in this group was 8.23 ±0.64%.
The control group (GC) consisted of 25 randomly selected healthy children who were treated at the NZOZ Pachońscy Dental Clinic in Tarnowskie Góry, Poland. The mean age of the subjects was 14.52 ±1.29 years. The group consisted of 13 girls and 12 boys.
The study groups were homogeneous in terms of sex and age (χ2 Pearson test: p = 0.50, p = 0.41). The study was conducted in the period 2014–2018.
Exclusion criteria from the research
The following groups were excluded: Patients and/or legal guardians, parents not agreeing to participate in the study, patients not willing to cooperate in the study, and those additionally burdened with other diseases (thyroid diseases, celiac disease, arterial hypertension).
In the control group, the exclusion criteria were diabetes and all systemic disorders, as well as refusal to participate in the study.
The tests were performed anonymously, and each patient was given a code number. The parents or legal guardians of all participants gave their written consent to participate in the study. The research project was approved by the Bioethical Commission of the Silesian Medical University in Katowice (No. KNW / 0022 / KB1 / 26 / I / 14 of 22/04/2014) and conducted in accordance with the latest version of the Helsinki declaration
Collection of the samples to microbiological tests
The material for microbiological tests was collected from each of the subjects as 2 swabs: one from the bottom of the oral cavity (BOC bacteria) and the other from the posterior part of the dorsum of the tongue (DT bacteria). The swabs were collected using sterile swabs, which were then placed in the transport medium and delivered to the Department of Microbiology and Immunology of the Medical University of Silesia in Zabrze, where the laboratory microbiological tests were carried out. The material was collected in the morning between 08:00 and 10:00 a.m. The participants were asked to attend the examination with an empty stomach and before completing any hygienic procedures in the oral cavity.
Laboratory microbiological tests
Microbiological tests were carried out using classical methods routinely used in microbiological diagnostics. The material collected from the examined patients was cultured on appropriate culture media to multiply and then isolate pure microbial cultures. Aerobic bacteria were multiplied on solid medium Columbia agar with a 5% addition of sheep blood at 37°C. Anaerobic bacteria were multiplied on Schaedler K3 solid medium with a 5% addition of sheep blood at 37°C in an-aerobic conditions obtained using Biomerieux Genbag Anaer kits (Marcy l’Etoile, France). Candida fungi were multiplied and initially identified using chromogenic medium ChromID Candida from Biomerieux (Marcy l’Etoile, France).
After isolation and multiplication of the cultured strains of microorganisms, their species identification was carried out using the following sets of reagents: ENTEROtest 24 N, NEFERMtest 24 N, STREPTOtest 24, STAPHYtest 24, ANAEROtest 23, OXItest, PYRAtest, and the computer program TNW_lite 6.5 for species identification of Erba-Lachema microorganisms (Brno, Czech Republic). The following biochemical tests from Biomerieux (Marcy l’Etoile, France) were also used: Catalysis, Slidex Staph Kit, and API Candida. Execution, reading, and interpretation of the test results were performed in accordance with the recommendations of manufacturers of diagnostic reagent kits.
Statistical analysis
All statistical analyses were performed using the STATISTICA Version 13 program package (StatSoft, Tulsa, OK, USA), the SciPy module, and the Statsmodels Phyton module. All charts were prepared using Excel (Microsoft Office, Redmond, Washington, USA). Continuous variables between the groups were compared using the Kruskal-Wallis and Mann-Whitney U tests because the analysed variables were not normally distributed.
All statistical analyses were supplemented with the results of 2 post-hoc tests: the Dunn-Bonferroni and Conover tests. To test the differences of nominal variables between the groups, the Pearson chi-square test was used. P values <0.05 were considered statistically significant.
Results
The results are presented in the form of box plots. The upper and lower boundaries of the box determine the first (Q1) and third quartiles (Q3) of the variable distribution, respectively. The line in the centre of the box is the median. Lower and upper whisker indicate the minimum and maximum values. The cultures were assessed according to the number of microbial species. The number of strains of a given species of microorganisms was determined. The authors counted the number of different microorganism strains that were isolated from mucosa of each particular subject per participant for the 3 mentioned groups of children: PC, WC, and GC.
Figure 1 presents the results of the Kruskal-Wallis test used to assess the presence of any significant differences in the number of all bacteria isolated from the bottom of the oral cavity (Bacteria BOC) between all study groups. These differences were shown to be statistically significant (p < 0.001), and as such the results of the statistical analysis were supported by 2 of the post-hoc tests: the Dunn-Bonferroni test and the Conover test. Post-hoc analysis revealed that the median number of different strains isolated by BOC swab was significantly lower in the GC group compared to the PC (p = 0.001) and WC groups (p < 0.001) (Table I). The obtained median in PC group was 4.0, 5.0 in WC group, and 3.0 in GC group. The highest number of isolated strains of bacteria [8] in one participant was found in the PC and GC groups. In the WC group this value was 7.
Figure 1
The number of bacterial strains isolated from the bottom of the oral cavity in the study groups of children (Kruskal- Wallis test)
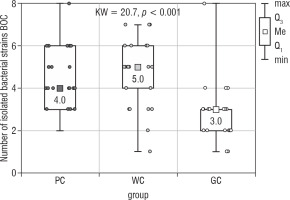
Table I
Results of post hoc tests – number of isolated bacterial strains (BOC)
| Post hoc | Dunn-Bonferroni | |||
|---|---|---|---|---|
| p | PC | WC | GC | |
| Conover | PC | 0.623 | 0.001 | |
| WC | 0.222 | < 0.001 | ||
| GC | 0.028 | 0.019 | ||
The number of all isolated bacteria from the tongue’s dorsum (Bacteria DT) also differed significantly between the examined groups of children (the Kruskal-Wallis test; p = 0.001; Fig. 2). The results of the Dunn-Bonferroni test showed statistically significant differences between the WC (median 3.0) and GC (median 3.0) groups as well as between the PC (median 4.0) and GC groups. The second post-hoc test revealed the statistically significant difference only between PC and GC groups (Table II). There were no statistical differences between the studied groups of children with diabetes.
Figure 2
The number of bacterial strains isolated from the dorsum of the tongue in the study groups of children (Kruskal-Wallis test)
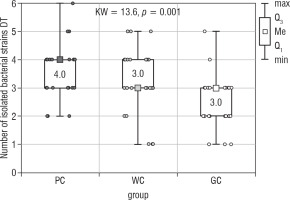
Table II
Results of post hoc tests – number of isolated bacterial strains (DT)
| Post hoc | Dunn-Bonferroni | |||
|---|---|---|---|---|
| p | PC | WC | GC | |
| Conover | PC | 0.368 | 0.001 | |
| WC | 0.166 | 0.021 | ||
| GC | 0.029 | 0.058 | ||
The conducted statistical analysis revealed statistically significant differences in the total number of all isolated microorganisms (Bacteria BOC & DT) between PC and GC groups (Kruskal Wallis test; p < 0.001; Fig. 3). The highest number of isolated bacterial strains [10] was obtained in the PC group. In the WC and GC groups the value was 9. The medians obtained in the PC, WC, and GC groups were: 6, 7, and 4, respectively. There were no statistically significant differences in the number of isolated microorganisms between the PC and WC groups (Table III).
Figure 3
The number of bacterial strains isolated from the bottom of the oral cavity and the dorsum of the tongue in the study groups of children (Kruskal-Wallis test)
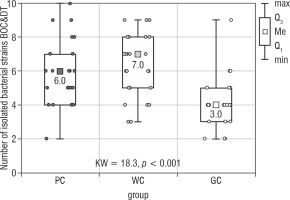
Table III
Results of post hoc tests – number of isolated bacterial strains (BOC&DT)
| Post hoc | Dunn-Bonferroni | |||
|---|---|---|---|---|
| p | PC | WC | GC | |
| Conover | PC | 0.507 | 0.003 | |
| WC | 0.195 | < 0.001 | ||
| GC | 0.035 | 0.022 | ||
As a result of the microbiological examination, the number of total microorganisms isolated in the PC group was 160 species. In the WC group, 158 microorganisms were cultured, and in the control group 103.
The microorganisms cultured were assigned to the following groups: gram-positive streptococci, gram-negative streptococci, gram-positive coccobacillus, and gram-negative coccobacillus (Table IV).
Table IV
Species of oral microbiota isolated from studied groups of children
A separate group isolated during the microbiological examination was the yeast Candida albicans. In the PC group, Candida albicans was isolated in 8 children, in the WC group in 4 diabetic children, and in the GC group in 5 healthy children. The obtained results were not statistically significant (p = 0.21; Table IV).
The largest group of isolated microorganisms comprised bacteria from the Streptococcus genus. As a result of the study, the following streptococci were cultured: S treptococcus acidominimus, S. cristatus, S. mitis, S. oralis, S. pneumoniae, S. pyogenes, S. sanguinis, and S. salivarius. Statistical calculations demonstrated that the total number of Streptococcus differed significantly between the studied groups (p = 0.018; Fig. 4). Both of the post hoc tests showed a significant difference between the WC and GC groups (Table V).
Figure 4
The number of Streptococcus strain isolated from the bottom of the oral cavity and the dorsum of the tongue in the study groups of children (Kruskal-Wallis test)
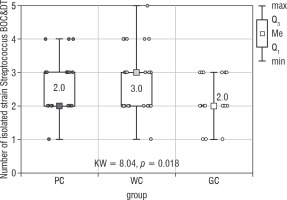
Table V
Results of post hoc tests – number of Streptococcus strain (BOC&DT)
| Post hoc | Dunn-Bonferroni | |||
|---|---|---|---|---|
| p | PC | WC | GC | |
| Conover | PC | 0.242 | 0.228 | |
| WC | 0.140 | 0.007 | ||
| GC | 0.137 | 0.049 | ||
Statistical analysis was also performed using the Mann-Whitney test between the combined groups of children with diabetes (PC & WC = GD) and the control group. This analysis showed a statistically significantly higher number of bacterial strains isolated from the bottom of the oral cavity and the dorsum of the tongue in the group of children with diabetes (GD) compared to the control group (Mann-Whitney U test; p < 0.001). Similar data were obtained with the Streptococcus strain. The GD group was characterized by a significantly higher number of these bacteria as compared to the control group (Mann-Whitney U test; p = 0.014; Figs. 5 and 6).
Discussion
Caries is the most common disease of the oral cavity to which humans are susceptible during their lifetime. This disease not only results in tooth destruction but also affects the pulp and periapical tissues. The most pathogenic bacteria related to the development of caries is Streptococcus mutans. Nevertheless, other bacteria including Streptococcus, Veillonella, Actinomyces, Granulicatella, Leptotrichia, Thiomonas, Bifidobacterium, and Prevotella species were detected in cases of S-ECC [16, 17]. A significantly higher number of bacteria from the Streptococcus genus were found in the group of children with well-controlled diabetes mellitus compared to healthy children. The samples that were taken from the floor of the oral cavity also revealed differences in the number of described bacteria between both examined WC and PC groups of diabetics. Streptococcus mitis, S. salivarius, S. sanguinis, and S. acidominimus were the most abundantly represented. The study of Janem et al. showed that the microbiological salivary profile of diabetics did not differ statistically significantly from healthy people [18]. Nevertheless, Kampoo et al. noticed distinctly higher levels of Streptococcus and Lactobacillus in diabetics with caries [19].
Our own research also noted an increased amount of Streptococcus mitis in the group of children with type 1 diabetes in comparison to healthy children. This bacterium was isolated in 88% of children in the PC group, 92% in the WC group, and 64% in the control group. Streptococcus mitis is classified as “low pH streptococcus” – this microorganism can reduce the pH of glucose broth to below 4.4. Species with similar abilities include S. oralis, S. anginosus, and S. gordonii. The organisms mentioned above are considered to have low carious potential and are listed amongst the primary colonizers of the pellicle. However, their capability of decreasing the pH level and therefore the occurrence of caries is confirmed by numerous studies [20, 21].
Candida spp. are commensals detected in the oral cavity among healthy individuals. However, their occurrence and virulence are significantly increased in immunocompromised patients. They may become pathogenic in the case of physiological changes in the host and cause oral candidiasis or invasive systemic infection [22]. Numerous studies identified an increased incidence of Candida spp. in type 1 diabetes, which was closely related to diabetes duration and level of decompensation [23]. Soysa et al. demonstrated that the incidence of that fungus on mucosa may concern 80% of subjects [24]. The study conducted by the authors did not reveal statistically significant differences in the number of isolated Candida spp. (p = 0.21). In the PC group Candida spp. were detected in 8 cases, in the WC group in 6 cases, and in the GC group in 5 cases. It is certainly related to the fact that the study group consisted of children aged 10–18 years with no periodontitis [25]. Candida albicans strains are often isolated in cases of co-existing diabetes and periodontitis. Sardi et al. revealed that the incidence of Candida albicans reaches 63.63% among patients with diabetes mellitus and chronic periodontitis [22].
Hintao et al. showed an increased incidence of Treponema denticola, Streptococcus sanguinis, Prevotella nigrescens, Staphylococcus intermedius, and Streptococcus oralis in the subgingival plaque of people with type 2 diabetes compared to healthy people, despite the lack of statistically significant differences in the examined subgingival plaque samples [26].
A similar composition of bacterial flora was observed among patients with type 1 DM and in the follow-up of non-diabetic diabetics with periodontitis [27].
Limitations of the study
The performed research relates to an interesting issue. However, we may be limited by an insufficient number of examined patients. On the other hand, the research was carried out on patients of the largest Diabetology Clinic in Upper Silesia, and applying the inclusion and exclusion criteria also significantly reduced the number of children who could take part in the research. Conducting the study in other regions of Poland to examine the possible influence of environmental factors on oral microbiota would be informative.

 POLSKI
POLSKI






